Starting Tiny with Protein LLaMA
Last month I posted an embedding model for proteins. With a new dataset of plant proteins (GreenBeing), I'm exploring training a set of models on a mix of proteins and natural language text. Bioinformatics research often looks at proteins' function, their location within the cell, and location within the organism. Combining with larger natural language models might allow Q&A, chat, descriptions of new proteins, and interpretation.
tinyllama-mixpretrain-quinoa-sciphi
This is a 1.1B TinyLLaMA model where I used Trainer to continue training on thousands of quinoa proteins and snippets of scientific text (SciPhi/textbooks-are-all-you-need-lite). Though I have considered adding 'biotokens' to exclusively represent amino acids, I haven't tried it yet. I shared the CoLab notebook from training.
Today I tried PEFT to finetune this model on proteins from other plants in the GreenBeing finetuning split. I'd like to DSPy this / make a Q&A format in the future, but for now it's:
<MMNPDGGDGDR…> function\nlocation\nother annotations
I left out proteins from the Zea genus (maize/corn) so I could evaluate the model. Links for Notebook and final LoRA.
If this project shows meaningful results, I will try training with a larger model and dataset (LLaMA 3 or Mixtral, the full pretraining split of GreenBeing). I'm not sure yet if the PEFT features for new tokens would be sufficient for biotokens. Also for some time I tried MergeKit on a protein-only TinyLLaMA and Stanford's BioMedLM, or a 16-bit version of PharMolix/BioMedGPT-LM-7B. The architectures and parameters didn't line up. Any tips for that, please let me know. 🙏
The GreenBeing Dataset in detail
GreenBeing is a dataset of proteins from food crops and their wild relatives. I decided to organize the dataset like this:
- pretraining split is amino acid sequences from select crop species and wild relatives found on UniProt
- finetuning split is reviewed sequences from the same taxa, plus text annotations from UniProtKB (knowledge base)
- research split is 99% quinoa, with some kaniwa and amaranth
Understanding UniProt Data
Most rice that you find at the grocery store is Oryza sativa. The japonica subspecies has ~4,100 proteins in the reviewed SwissProt dataset, and ~44,700 in unreviewed TrEMBL. This is more than the number of genes which Google and UniProt say that rice has, so I don't know if it's duplicates, variants, or errors. From reading up on what "unreviewed" means, I'm satisfied to use these sequences in pretraining so long as we don't use any automated / predicted annotations.
The sequences use IUPAC-IUB codes where letters A-Z map to amino acids. This is different from genomic data which would use nucleotides (ACTG) and models could cover gene expression.
NOTE: When searching UniProt, use taxonomy_name:__, plus taxonomy_name:Viridiplantae, so you see Zea, Triticeae, etc. and not an all-text search which includes their viruses and pests!

Pretraining Split
I moved up rice's taxonomy tree to Oryzeae to include wild rice, and I've found satisfying levels for other popular crops (corn, soybean, wheat, tomato). There's a species column in the dataset for filtering, but there's interesting research about fortifying and diversifying crops with genes from their wild relatives.
Here's a distribution of proteins in this set, with Papilionoideae (soybeans, peas, chickpeas, peanuts, etc) taking the lead:
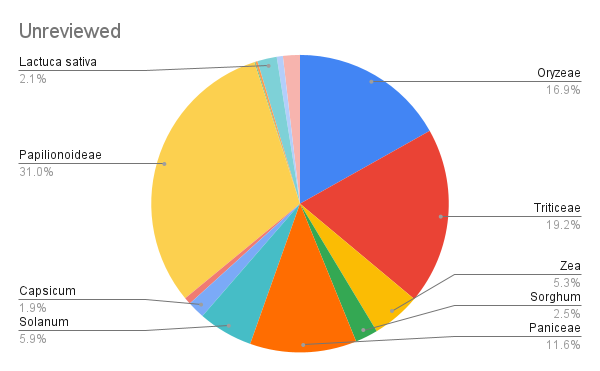
Finetuning Split
I used the same taxa to query reviewed/SwissProt proteins. Rice has excellent coverage in the set compared to other plants - just under half of the finetuning split.

Research Split
UniProt has a near-complete quinoa proteome but their status is unreviewed - even the genes that get you red/white/black seeds are still being identified. You could use protein embeddings to match these to the closest reviewed proteins in other species. I'm hopeful this can contribute to quinoa research as it's being considered in many regions as a drought-resistant crop.
Limitations and Safety Notes
UniProt proteins downloaded March 29, 2024.
There could be significant overlap between the reviewed and unreviewed proteins from similar species and accessions.
Species include inedible wild relatives.
Many people have allergic reactions to wheat/gluten, nightshades, maize, and other crops.
Chili peppers can be spicy. 🌶️