Datasets:
Tasks:
Image Segmentation
Modalities:
Image
Sub-tasks:
semantic-segmentation
Languages:
English
Size:
10K - 100K
License:
Update README.md
Browse files
README.md
CHANGED
|
@@ -21,3 +21,95 @@ task_categories:
|
|
| 21 |
task_ids:
|
| 22 |
- semantic-segmentation
|
| 23 |
---
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 21 |
task_ids:
|
| 22 |
- semantic-segmentation
|
| 23 |
---
|
| 24 |
+
|
| 25 |
+
|
| 26 |
+
## Setup
|
| 27 |
+
|
| 28 |
+
```bash
|
| 29 |
+
pip install datasets
|
| 30 |
+
```
|
| 31 |
+
|
| 32 |
+
## Feel the Magic
|
| 33 |
+
|
| 34 |
+
### Load Dataset
|
| 35 |
+
```python
|
| 36 |
+
from datasets import load_dataset
|
| 37 |
+
|
| 38 |
+
data = load_dataset('ziq/RSNA-ATD2023')
|
| 39 |
+
print(data)
|
| 40 |
+
```
|
| 41 |
+
|
| 42 |
+
```bash
|
| 43 |
+
DatasetDict({
|
| 44 |
+
train: Dataset({
|
| 45 |
+
features: ['patient_id', 'series_id', 'frame_id', 'image', 'mask'],
|
| 46 |
+
num_rows: 70291
|
| 47 |
+
})
|
| 48 |
+
})
|
| 49 |
+
```
|
| 50 |
+
|
| 51 |
+
### Train Test Split
|
| 52 |
+
```python
|
| 53 |
+
data = data['train'].train_test_split(test_size=0.2)
|
| 54 |
+
```
|
| 55 |
+
|
| 56 |
+
```python
|
| 57 |
+
train, test = data['train'], data['test']
|
| 58 |
+
|
| 59 |
+
# train[0]['patient_id']
|
| 60 |
+
# train[0]['image'] -> PIL Image
|
| 61 |
+
# train[0]['mask'] -> PIL Image
|
| 62 |
+
```
|
| 63 |
+
|
| 64 |
+
### Get Segmentation Mask
|
| 65 |
+
|
| 66 |
+
```python
|
| 67 |
+
ids = 3
|
| 68 |
+
image, mask = train[ids]['image'], train[ids]['mask']
|
| 69 |
+
|
| 70 |
+
mask = np.array(mask)
|
| 71 |
+
liver, spleen, right_kidney, left_kidney, bowel = [(mask == i,1,0)[0] * i for i in range(1, len(labels))]
|
| 72 |
+
```
|
| 73 |
+
|
| 74 |
+
|
| 75 |
+
### Plot Mask
|
| 76 |
+
|
| 77 |
+
```python
|
| 78 |
+
from matplotlib.colors import ListedColormap, BoundaryNorm
|
| 79 |
+
|
| 80 |
+
colors = ['#02020e', '#520e6d', '#c13a50', '#f57d15', '#fac62c', '#f4f88e'] # inferno
|
| 81 |
+
bounds = range(0, len(colors) + 1)
|
| 82 |
+
|
| 83 |
+
# Define the boundaries for each class in the colormap
|
| 84 |
+
cmap, norm = ListedColormap(colors), BoundaryNorm(bounds, len(colors))
|
| 85 |
+
|
| 86 |
+
# Plot the segmentation mask with the custom colormap
|
| 87 |
+
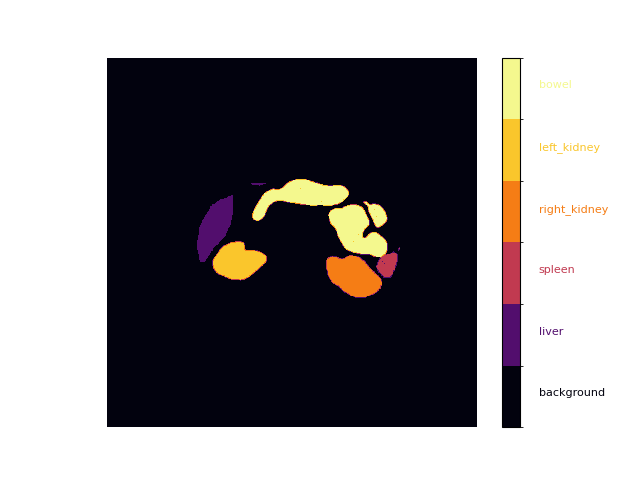
def plot_mask(mask, alpha=1.0):
|
| 88 |
+
_, ax = plt.subplots()
|
| 89 |
+
cax = ax.imshow(mask, cmap=cmap, norm=norm, alpha=alpha)
|
| 90 |
+
cbar = plt.colorbar(cax, cmap=cmap, norm=norm, boundaries=bounds, ticks=bounds)
|
| 91 |
+
cbar.set_ticks([])
|
| 92 |
+
_labels = [""] + labels
|
| 93 |
+
for i in range(1, len(_labels)):
|
| 94 |
+
cbar.ax.text(2, -0.5 + i, _labels[i], ha='left', color=colors[i - 1], fontsize=8)
|
| 95 |
+
plt.axis('off')
|
| 96 |
+
plt.show()
|
| 97 |
+
|
| 98 |
+
def plot(image, mask, cmap='gray'):
|
| 99 |
+
plt.imshow(image * np.where(mask > 0,1,0), cmap=cmap)
|
| 100 |
+
plt.axis('off')
|
| 101 |
+
plt.show()
|
| 102 |
+
```
|
| 103 |
+
|
| 104 |
+
```python
|
| 105 |
+
plot(image, mask)
|
| 106 |
+
```
|
| 107 |
+
|
| 108 |
+

|
| 109 |
+
|
| 110 |
+
```python
|
| 111 |
+
plot_mask(mask)
|
| 112 |
+
```
|
| 113 |
+
|
| 114 |
+
|
| 115 |
+
|