fixed some small changes with formatting in readme
Browse files- README.md +29 -32
- configs/metadata.json +2 -1
- docs/README.md +29 -32
README.md
CHANGED
|
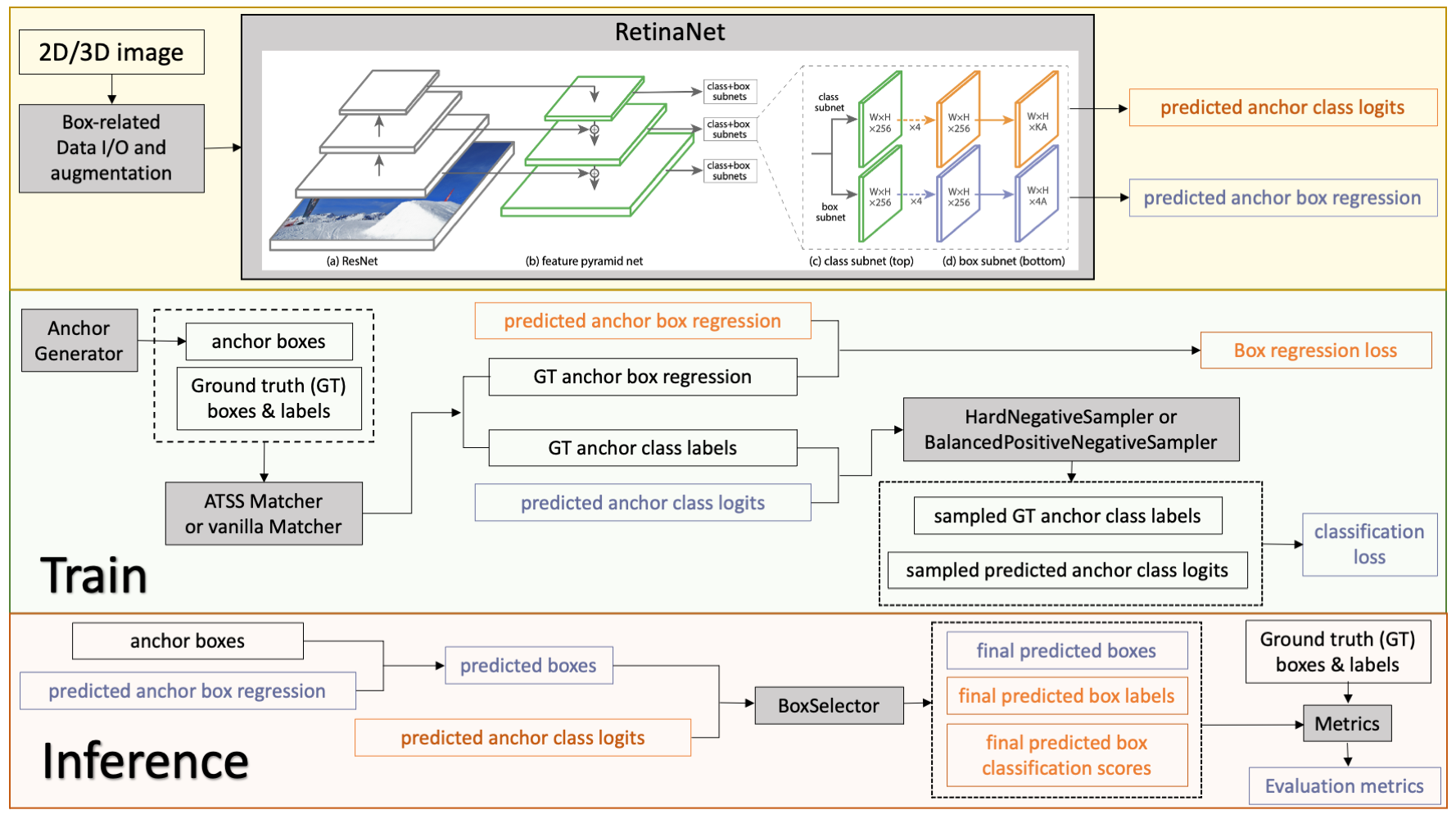
@@ -12,76 +12,77 @@ This model is trained on LUNA16 dataset (https://luna16.grand-challenge.org/Home
|
|
| 12 |
|
| 13 |
|
| 14 |
|
| 15 |
-
##
|
| 16 |
-
### 1.1 Data description
|
| 17 |
The dataset we are experimenting in this example is LUNA16 (https://luna16.grand-challenge.org/Home/), which is based on [LIDC-IDRI database](https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) [3,4,5].
|
| 18 |
|
| 19 |
LUNA16 is a public dataset of CT lung nodule detection. Using raw CT scans, the goal is to identify locations of possible nodules, and to assign a probability for being a nodule to each location.
|
| 20 |
|
| 21 |
Disclaimer: We are not the host of the data. Please make sure to read the requirements and usage policies of the data and give credit to the authors of the dataset! We acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study.
|
| 22 |
|
| 23 |
-
###
|
| 24 |
We follow the official 10-fold data splitting from LUNA16 challenge and generate data split json files using the script from [nnDetection](https://github.com/MIC-DKFZ/nnDetection/blob/main/projects/Task016_Luna/scripts/prepare.py).
|
| 25 |
|
| 26 |
Please download the resulted json files from https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/LUNA16_datasplit-20220615T233840Z-001.zip.
|
| 27 |
|
| 28 |
In these files, the values of "box" are the ground truth boxes in world coordinate.
|
| 29 |
|
| 30 |
-
###
|
| 31 |
The raw CT images in LUNA16 have various of voxel sizes. The first step is to resample them to the same voxel size.
|
| 32 |
In this model, we resampled them into 0.703125 x 0.703125 x 1.25 mm.
|
| 33 |
|
| 34 |
Please following the instruction in Section 3.1 of https://github.com/Project-MONAI/tutorials/tree/main/detection to do the resampling.
|
| 35 |
|
| 36 |
-
###
|
| 37 |
The mhd/raw original data can be downloaded from [LUNA16](https://luna16.grand-challenge.org/Home/). The DICOM original data can be downloaded from [LIDC-IDRI database](https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) [3,4,5]. You will need to resample the original data to start training.
|
| 38 |
|
| 39 |
Alternatively, we provide [resampled nifti images](https://drive.google.com/drive/folders/1JozrufA1VIZWJIc5A1EMV3J4CNCYovKK?usp=share_link) and a copy of [original mhd/raw images](https://drive.google.com/drive/folders/1-enN4eNEnKmjltevKg3W2V-Aj0nriQWE?usp=share_link) from [LUNA16](https://luna16.grand-challenge.org/Home/) for users to download.
|
| 40 |
|
| 41 |
-
##
|
| 42 |
-
The training was the following:
|
| 43 |
|
| 44 |
-
GPU: at least 16GB GPU memory
|
| 45 |
-
|
| 46 |
-
|
| 47 |
-
|
| 48 |
-
|
| 49 |
-
|
| 50 |
-
Optimizer: Adam
|
| 51 |
-
|
| 52 |
-
Learning Rate: 1e-2
|
| 53 |
-
|
| 54 |
-
Loss: BCE loss and L1 loss
|
| 55 |
|
| 56 |
### Input
|
| 57 |
-
|
|
|
|
| 58 |
|
| 59 |
### Output
|
| 60 |
-
In
|
| 61 |
|
| 62 |
-
In
|
| 63 |
|
| 64 |
-
##
|
| 65 |
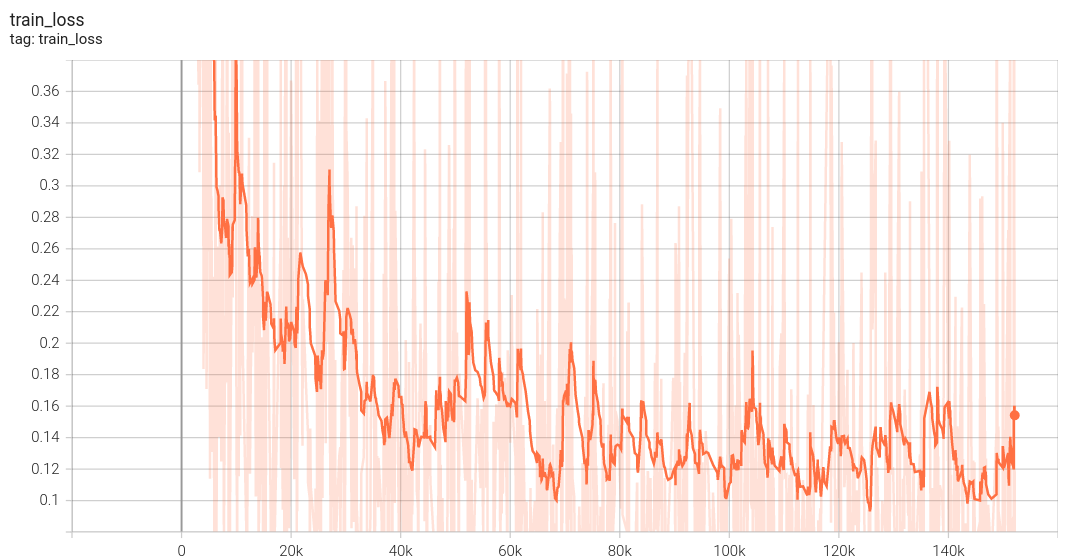
Coco metric is used for evaluating the performance of the model. The pre-trained model was trained and validated on data fold 0. This model achieves a mAP=0.853, mAR=0.994, AP(IoU=0.1)=0.862, AR(IoU=0.1)=1.0.
|
| 66 |
|
| 67 |
-
|
|
|
|
| 68 |
|
|
|
|
| 69 |
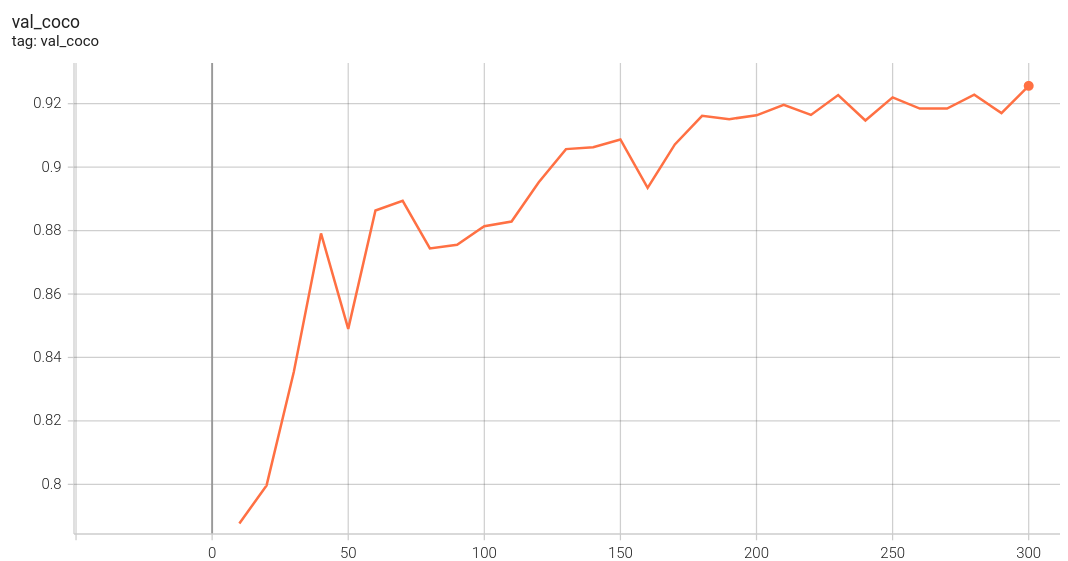
The validation accuracy in this curve is the mean of mAP, mAR, AP(IoU=0.1), and AR(IoU=0.1) in Coco metric.
|
| 70 |
|
| 71 |
-
|
| 72 |
|
| 73 |
-
##
|
| 74 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
| 75 |
```
|
| 76 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 77 |
```
|
| 78 |
|
| 79 |
-
Override the `train` config to execute evaluation with the trained model:
|
| 80 |
```
|
| 81 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 82 |
```
|
| 83 |
|
| 84 |
-
Execute inference on resampled LUNA16 images by setting `"whether_raw_luna16": false` in `inference.json`:
|
| 85 |
```
|
| 86 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 87 |
```
|
|
@@ -91,10 +92,6 @@ Note that in inference.json, the transform "LoadImaged" in "preprocessing" and "
|
|
| 91 |
This depends on the input images. LUNA16 needs `"affine_lps_to_ras": true`.
|
| 92 |
It is possible that your inference dataset should set `"affine_lps_to_ras": false`.
|
| 93 |
|
| 94 |
-
|
| 95 |
-
# Disclaimer
|
| 96 |
-
This is an example, not to be used for diagnostic purposes.
|
| 97 |
-
|
| 98 |
# References
|
| 99 |
[1] Lin, Tsung-Yi, et al. "Focal loss for dense object detection." ICCV 2017. https://arxiv.org/abs/1708.02002)
|
| 100 |
|
|
|
|
| 12 |
|
| 13 |

|
| 14 |
|
| 15 |
+
## Data
|
|
|
|
| 16 |
The dataset we are experimenting in this example is LUNA16 (https://luna16.grand-challenge.org/Home/), which is based on [LIDC-IDRI database](https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) [3,4,5].
|
| 17 |
|
| 18 |
LUNA16 is a public dataset of CT lung nodule detection. Using raw CT scans, the goal is to identify locations of possible nodules, and to assign a probability for being a nodule to each location.
|
| 19 |
|
| 20 |
Disclaimer: We are not the host of the data. Please make sure to read the requirements and usage policies of the data and give credit to the authors of the dataset! We acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study.
|
| 21 |
|
| 22 |
+
### 10-fold data splitting
|
| 23 |
We follow the official 10-fold data splitting from LUNA16 challenge and generate data split json files using the script from [nnDetection](https://github.com/MIC-DKFZ/nnDetection/blob/main/projects/Task016_Luna/scripts/prepare.py).
|
| 24 |
|
| 25 |
Please download the resulted json files from https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/LUNA16_datasplit-20220615T233840Z-001.zip.
|
| 26 |
|
| 27 |
In these files, the values of "box" are the ground truth boxes in world coordinate.
|
| 28 |
|
| 29 |
+
### Data resampling
|
| 30 |
The raw CT images in LUNA16 have various of voxel sizes. The first step is to resample them to the same voxel size.
|
| 31 |
In this model, we resampled them into 0.703125 x 0.703125 x 1.25 mm.
|
| 32 |
|
| 33 |
Please following the instruction in Section 3.1 of https://github.com/Project-MONAI/tutorials/tree/main/detection to do the resampling.
|
| 34 |
|
| 35 |
+
### Data download
|
| 36 |
The mhd/raw original data can be downloaded from [LUNA16](https://luna16.grand-challenge.org/Home/). The DICOM original data can be downloaded from [LIDC-IDRI database](https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) [3,4,5]. You will need to resample the original data to start training.
|
| 37 |
|
| 38 |
Alternatively, we provide [resampled nifti images](https://drive.google.com/drive/folders/1JozrufA1VIZWJIc5A1EMV3J4CNCYovKK?usp=share_link) and a copy of [original mhd/raw images](https://drive.google.com/drive/folders/1-enN4eNEnKmjltevKg3W2V-Aj0nriQWE?usp=share_link) from [LUNA16](https://luna16.grand-challenge.org/Home/) for users to download.
|
| 39 |
|
| 40 |
+
## Training configuration
|
| 41 |
+
The training was performed with the following:
|
| 42 |
|
| 43 |
+
- GPU: at least 16GB GPU memory
|
| 44 |
+
- Actual Model Input: 192 x 192 x 80
|
| 45 |
+
- AMP: True
|
| 46 |
+
- Optimizer: Adam
|
| 47 |
+
- Learning Rate: 1e-2
|
| 48 |
+
- Loss: BCE loss and L1 loss
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 49 |
|
| 50 |
### Input
|
| 51 |
+
1 channel
|
| 52 |
+
- List of 3D CT patches
|
| 53 |
|
| 54 |
### Output
|
| 55 |
+
In Training Mode: A dictionary of classification and box regression loss.
|
| 56 |
|
| 57 |
+
In Evaluation Mode: A list of dictionaries of predicted box, classification label, and classification score.
|
| 58 |
|
| 59 |
+
## Performance
|
| 60 |
Coco metric is used for evaluating the performance of the model. The pre-trained model was trained and validated on data fold 0. This model achieves a mAP=0.853, mAR=0.994, AP(IoU=0.1)=0.862, AR(IoU=0.1)=1.0.
|
| 61 |
|
| 62 |
+
#### Training Loss
|
| 63 |
+
|
| 64 |
|
| 65 |
+
#### Validation Accuracy
|
| 66 |
The validation accuracy in this curve is the mean of mAP, mAR, AP(IoU=0.1), and AR(IoU=0.1) in Coco metric.
|
| 67 |
|
| 68 |
+

|
| 69 |
|
| 70 |
+
## MONAI Bundle Commands
|
| 71 |
+
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
|
| 72 |
+
|
| 73 |
+
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
|
| 74 |
+
|
| 75 |
+
#### Execute training:
|
| 76 |
```
|
| 77 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 78 |
```
|
| 79 |
|
| 80 |
+
#### Override the `train` config to execute evaluation with the trained model:
|
| 81 |
```
|
| 82 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 83 |
```
|
| 84 |
|
| 85 |
+
#### Execute inference on resampled LUNA16 images by setting `"whether_raw_luna16": false` in `inference.json`:
|
| 86 |
```
|
| 87 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 88 |
```
|
|
|
|
| 92 |
This depends on the input images. LUNA16 needs `"affine_lps_to_ras": true`.
|
| 93 |
It is possible that your inference dataset should set `"affine_lps_to_ras": false`.
|
| 94 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 95 |
# References
|
| 96 |
[1] Lin, Tsung-Yi, et al. "Focal loss for dense object detection." ICCV 2017. https://arxiv.org/abs/1708.02002)
|
| 97 |
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.4.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.4.4": "add data resource to readme",
|
| 6 |
"0.4.3": "update val patch size to avoid warning in monai 1.0.1",
|
| 7 |
"0.4.2": "update to use monai 1.0.1",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.4.5",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.4.5": "fixed some small changes with formatting in readme",
|
| 6 |
"0.4.4": "add data resource to readme",
|
| 7 |
"0.4.3": "update val patch size to avoid warning in monai 1.0.1",
|
| 8 |
"0.4.2": "update to use monai 1.0.1",
|
docs/README.md
CHANGED
|
@@ -5,76 +5,77 @@ This model is trained on LUNA16 dataset (https://luna16.grand-challenge.org/Home
|
|
| 5 |
|
| 6 |

|
| 7 |
|
| 8 |
-
##
|
| 9 |
-
### 1.1 Data description
|
| 10 |
The dataset we are experimenting in this example is LUNA16 (https://luna16.grand-challenge.org/Home/), which is based on [LIDC-IDRI database](https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) [3,4,5].
|
| 11 |
|
| 12 |
LUNA16 is a public dataset of CT lung nodule detection. Using raw CT scans, the goal is to identify locations of possible nodules, and to assign a probability for being a nodule to each location.
|
| 13 |
|
| 14 |
Disclaimer: We are not the host of the data. Please make sure to read the requirements and usage policies of the data and give credit to the authors of the dataset! We acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study.
|
| 15 |
|
| 16 |
-
###
|
| 17 |
We follow the official 10-fold data splitting from LUNA16 challenge and generate data split json files using the script from [nnDetection](https://github.com/MIC-DKFZ/nnDetection/blob/main/projects/Task016_Luna/scripts/prepare.py).
|
| 18 |
|
| 19 |
Please download the resulted json files from https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/LUNA16_datasplit-20220615T233840Z-001.zip.
|
| 20 |
|
| 21 |
In these files, the values of "box" are the ground truth boxes in world coordinate.
|
| 22 |
|
| 23 |
-
###
|
| 24 |
The raw CT images in LUNA16 have various of voxel sizes. The first step is to resample them to the same voxel size.
|
| 25 |
In this model, we resampled them into 0.703125 x 0.703125 x 1.25 mm.
|
| 26 |
|
| 27 |
Please following the instruction in Section 3.1 of https://github.com/Project-MONAI/tutorials/tree/main/detection to do the resampling.
|
| 28 |
|
| 29 |
-
###
|
| 30 |
The mhd/raw original data can be downloaded from [LUNA16](https://luna16.grand-challenge.org/Home/). The DICOM original data can be downloaded from [LIDC-IDRI database](https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) [3,4,5]. You will need to resample the original data to start training.
|
| 31 |
|
| 32 |
Alternatively, we provide [resampled nifti images](https://drive.google.com/drive/folders/1JozrufA1VIZWJIc5A1EMV3J4CNCYovKK?usp=share_link) and a copy of [original mhd/raw images](https://drive.google.com/drive/folders/1-enN4eNEnKmjltevKg3W2V-Aj0nriQWE?usp=share_link) from [LUNA16](https://luna16.grand-challenge.org/Home/) for users to download.
|
| 33 |
|
| 34 |
-
##
|
| 35 |
-
The training was the following:
|
| 36 |
|
| 37 |
-
GPU: at least 16GB GPU memory
|
| 38 |
-
|
| 39 |
-
|
| 40 |
-
|
| 41 |
-
|
| 42 |
-
|
| 43 |
-
Optimizer: Adam
|
| 44 |
-
|
| 45 |
-
Learning Rate: 1e-2
|
| 46 |
-
|
| 47 |
-
Loss: BCE loss and L1 loss
|
| 48 |
|
| 49 |
### Input
|
| 50 |
-
|
|
|
|
| 51 |
|
| 52 |
### Output
|
| 53 |
-
In
|
| 54 |
|
| 55 |
-
In
|
| 56 |
|
| 57 |
-
##
|
| 58 |
Coco metric is used for evaluating the performance of the model. The pre-trained model was trained and validated on data fold 0. This model achieves a mAP=0.853, mAR=0.994, AP(IoU=0.1)=0.862, AR(IoU=0.1)=1.0.
|
| 59 |
|
| 60 |
-
|
|
|
|
| 61 |
|
|
|
|
| 62 |
The validation accuracy in this curve is the mean of mAP, mAR, AP(IoU=0.1), and AR(IoU=0.1) in Coco metric.
|
| 63 |
|
| 64 |
-

|
| 65 |
|
| 66 |
-
##
|
| 67 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
| 68 |
```
|
| 69 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 70 |
```
|
| 71 |
|
| 72 |
-
Override the `train` config to execute evaluation with the trained model:
|
| 73 |
```
|
| 74 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 75 |
```
|
| 76 |
|
| 77 |
-
Execute inference on resampled LUNA16 images by setting `"whether_raw_luna16": false` in `inference.json`:
|
| 78 |
```
|
| 79 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 80 |
```
|
|
@@ -84,10 +85,6 @@ Note that in inference.json, the transform "LoadImaged" in "preprocessing" and "
|
|
| 84 |
This depends on the input images. LUNA16 needs `"affine_lps_to_ras": true`.
|
| 85 |
It is possible that your inference dataset should set `"affine_lps_to_ras": false`.
|
| 86 |
|
| 87 |
-
|
| 88 |
-
# Disclaimer
|
| 89 |
-
This is an example, not to be used for diagnostic purposes.
|
| 90 |
-
|
| 91 |
# References
|
| 92 |
[1] Lin, Tsung-Yi, et al. "Focal loss for dense object detection." ICCV 2017. https://arxiv.org/abs/1708.02002)
|
| 93 |
|
|
|
|
| 5 |
|
| 6 |

|
| 7 |
|
| 8 |
+
## Data
|
|
|
|
| 9 |
The dataset we are experimenting in this example is LUNA16 (https://luna16.grand-challenge.org/Home/), which is based on [LIDC-IDRI database](https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) [3,4,5].
|
| 10 |
|
| 11 |
LUNA16 is a public dataset of CT lung nodule detection. Using raw CT scans, the goal is to identify locations of possible nodules, and to assign a probability for being a nodule to each location.
|
| 12 |
|
| 13 |
Disclaimer: We are not the host of the data. Please make sure to read the requirements and usage policies of the data and give credit to the authors of the dataset! We acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study.
|
| 14 |
|
| 15 |
+
### 10-fold data splitting
|
| 16 |
We follow the official 10-fold data splitting from LUNA16 challenge and generate data split json files using the script from [nnDetection](https://github.com/MIC-DKFZ/nnDetection/blob/main/projects/Task016_Luna/scripts/prepare.py).
|
| 17 |
|
| 18 |
Please download the resulted json files from https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/LUNA16_datasplit-20220615T233840Z-001.zip.
|
| 19 |
|
| 20 |
In these files, the values of "box" are the ground truth boxes in world coordinate.
|
| 21 |
|
| 22 |
+
### Data resampling
|
| 23 |
The raw CT images in LUNA16 have various of voxel sizes. The first step is to resample them to the same voxel size.
|
| 24 |
In this model, we resampled them into 0.703125 x 0.703125 x 1.25 mm.
|
| 25 |
|
| 26 |
Please following the instruction in Section 3.1 of https://github.com/Project-MONAI/tutorials/tree/main/detection to do the resampling.
|
| 27 |
|
| 28 |
+
### Data download
|
| 29 |
The mhd/raw original data can be downloaded from [LUNA16](https://luna16.grand-challenge.org/Home/). The DICOM original data can be downloaded from [LIDC-IDRI database](https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) [3,4,5]. You will need to resample the original data to start training.
|
| 30 |
|
| 31 |
Alternatively, we provide [resampled nifti images](https://drive.google.com/drive/folders/1JozrufA1VIZWJIc5A1EMV3J4CNCYovKK?usp=share_link) and a copy of [original mhd/raw images](https://drive.google.com/drive/folders/1-enN4eNEnKmjltevKg3W2V-Aj0nriQWE?usp=share_link) from [LUNA16](https://luna16.grand-challenge.org/Home/) for users to download.
|
| 32 |
|
| 33 |
+
## Training configuration
|
| 34 |
+
The training was performed with the following:
|
| 35 |
|
| 36 |
+
- GPU: at least 16GB GPU memory
|
| 37 |
+
- Actual Model Input: 192 x 192 x 80
|
| 38 |
+
- AMP: True
|
| 39 |
+
- Optimizer: Adam
|
| 40 |
+
- Learning Rate: 1e-2
|
| 41 |
+
- Loss: BCE loss and L1 loss
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 42 |
|
| 43 |
### Input
|
| 44 |
+
1 channel
|
| 45 |
+
- List of 3D CT patches
|
| 46 |
|
| 47 |
### Output
|
| 48 |
+
In Training Mode: A dictionary of classification and box regression loss.
|
| 49 |
|
| 50 |
+
In Evaluation Mode: A list of dictionaries of predicted box, classification label, and classification score.
|
| 51 |
|
| 52 |
+
## Performance
|
| 53 |
Coco metric is used for evaluating the performance of the model. The pre-trained model was trained and validated on data fold 0. This model achieves a mAP=0.853, mAR=0.994, AP(IoU=0.1)=0.862, AR(IoU=0.1)=1.0.
|
| 54 |
|
| 55 |
+
#### Training Loss
|
| 56 |
+

|
| 57 |
|
| 58 |
+
#### Validation Accuracy
|
| 59 |
The validation accuracy in this curve is the mean of mAP, mAR, AP(IoU=0.1), and AR(IoU=0.1) in Coco metric.
|
| 60 |
|
| 61 |
+

|
| 62 |
|
| 63 |
+
## MONAI Bundle Commands
|
| 64 |
+
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
|
| 65 |
+
|
| 66 |
+
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
|
| 67 |
+
|
| 68 |
+
#### Execute training:
|
| 69 |
```
|
| 70 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 71 |
```
|
| 72 |
|
| 73 |
+
#### Override the `train` config to execute evaluation with the trained model:
|
| 74 |
```
|
| 75 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 76 |
```
|
| 77 |
|
| 78 |
+
#### Execute inference on resampled LUNA16 images by setting `"whether_raw_luna16": false` in `inference.json`:
|
| 79 |
```
|
| 80 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 81 |
```
|
|
|
|
| 85 |
This depends on the input images. LUNA16 needs `"affine_lps_to_ras": true`.
|
| 86 |
It is possible that your inference dataset should set `"affine_lps_to_ras": false`.
|
| 87 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 88 |
# References
|
| 89 |
[1] Lin, Tsung-Yi, et al. "Focal loss for dense object detection." ICCV 2017. https://arxiv.org/abs/1708.02002)
|
| 90 |
|