restructure readme to match updated template
Browse files- README.md +20 -26
- configs/metadata.json +2 -1
- docs/README.md +20 -26
README.md
CHANGED
|
@@ -9,8 +9,6 @@ license: apache-2.0
|
|
| 9 |
|
| 10 |
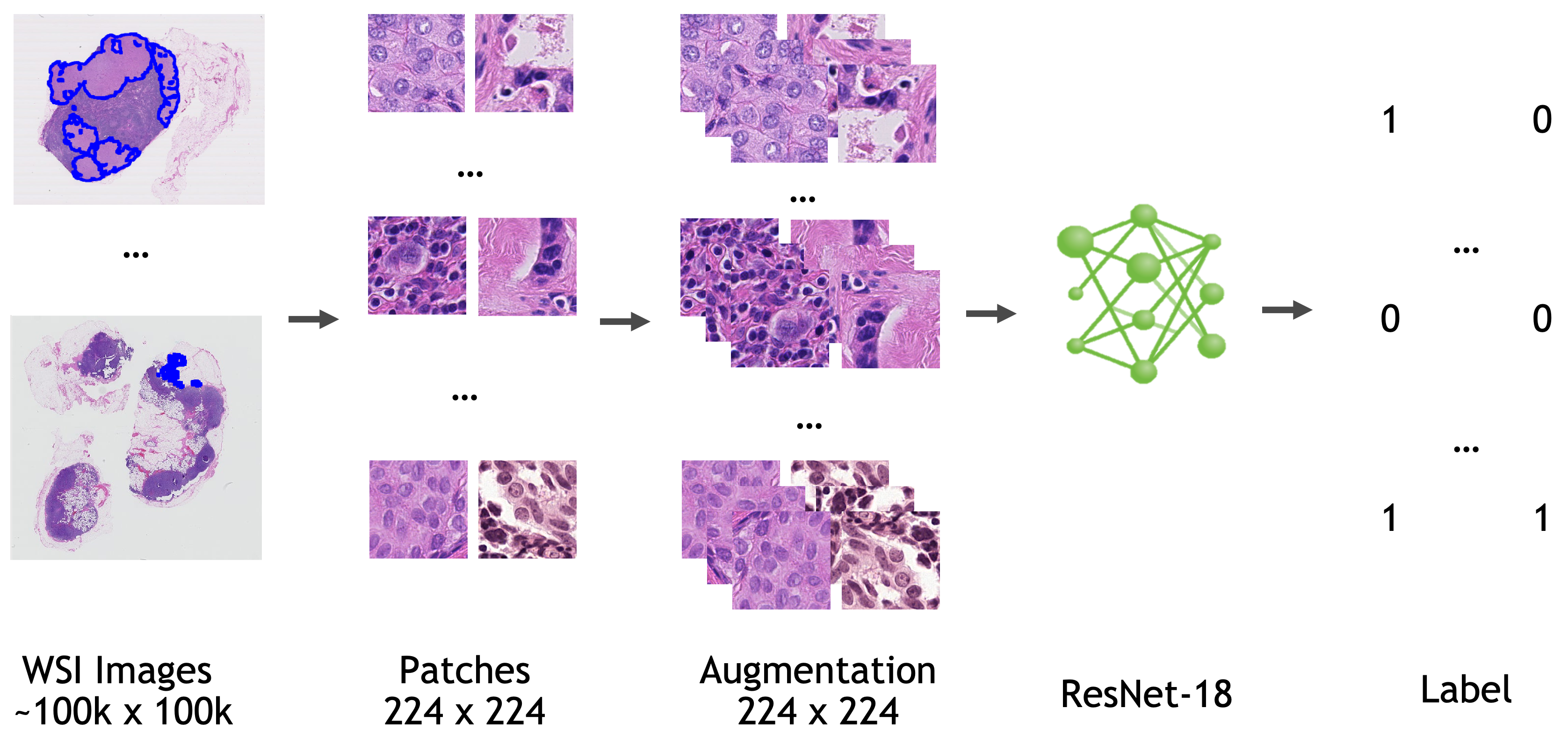
A pre-trained model for automated detection of metastases in whole-slide histopathology images.
|
| 11 |
|
| 12 |
-
## Workflow
|
| 13 |
-
|
| 14 |
The model is trained based on ResNet18 [1] with the last fully connected layer replaced by a 1x1 convolution layer.
|
| 15 |
|
| 16 |
|
|
@@ -27,7 +25,7 @@ Annotation information are adopted from [NCRF/jsons](https://github.com/baidu-re
|
|
| 27 |
- Modality: Histopathology
|
| 28 |
- Size: 270 WSIs for training/validation, 48 WSIs for testing
|
| 29 |
|
| 30 |
-
###
|
| 31 |
|
| 32 |
This bundle expects the training/validation data (whole slide images) reside in a `{data_root}/training/images`. By default `data_root` is pointing to `/workspace/data/medical/pathology/` You can modify `data_root` in the bundle config files to point to a different directory.
|
| 33 |
|
|
@@ -35,7 +33,7 @@ To reduce the computation burden during the inference, patches are extracted onl
|
|
| 35 |
|
| 36 |
Please refer to "Annotation" section of [Camelyon challenge](https://camelyon17.grand-challenge.org/Data/) to prepare ground truth images, which are needed for FROC computation. By default, this data set is expected to be at `/workspace/data/medical/pathology/ground_truths`. But it can be modified in `evaluate_froc.sh`.
|
| 37 |
|
| 38 |
-
|
| 39 |
|
| 40 |
The training was performed with the following:
|
| 41 |
|
|
@@ -48,61 +46,57 @@ The training was performed with the following:
|
|
| 48 |
- Loss: BCEWithLogitsLoss
|
| 49 |
- Whole slide image reader: cuCIM (if running on Windows or Mac, please install `OpenSlide` on your system and change `wsi_reader` to "OpenSlide")
|
| 50 |
|
| 51 |
-
|
| 52 |
-
|
| 53 |
-
Input: Input for the training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
| 54 |
|
| 55 |
-
|
| 56 |
-
2. Randomly applying color jittering
|
| 57 |
-
3. Randomly applying spatial flipping
|
| 58 |
-
4. Randomly applying spatial rotation
|
| 59 |
-
5. Randomly applying spatial zooming
|
| 60 |
-
6. Randomly applying intensity scaling
|
| 61 |
|
| 62 |
-
|
| 63 |
|
| 64 |
-
|
| 65 |
|
| 66 |
-
|
| 67 |
|
| 68 |
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 69 |
|
| 70 |
-
|
| 71 |
|
| 72 |
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 73 |
This model achieve the ~0.91 accuracy on validation patches, and FROC of 0.685 on the 48 Camelyon testing data that have ground truth annotations available.
|
| 74 |
-

|
| 75 |
|
| 76 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 77 |
|
| 78 |
-
Execute training:
|
| 79 |
|
| 80 |
```
|
| 81 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 82 |
```
|
| 83 |
|
| 84 |
-
Override the `train` config to execute multi-GPU training:
|
| 85 |
|
| 86 |
```
|
| 87 |
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 88 |
```
|
| 89 |
|
| 90 |
-
Please note that the distributed training
|
| 91 |
-
Please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html) for more details.
|
| 92 |
|
| 93 |
-
Execute inference:
|
| 94 |
|
| 95 |
```
|
| 96 |
CUDA_LAUNCH_BLOCKING=1 python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 97 |
```
|
| 98 |
|
| 99 |
-
Evaluate FROC metric:
|
| 100 |
|
| 101 |
```
|
| 102 |
cd scripts && source evaluate_froc.sh
|
| 103 |
```
|
| 104 |
|
| 105 |
-
Export checkpoint to TorchScript file:
|
| 106 |
|
| 107 |
TorchScript conversion is currently not supported.
|
| 108 |
|
|
|
|
| 9 |
|
| 10 |
A pre-trained model for automated detection of metastases in whole-slide histopathology images.
|
| 11 |
|
|
|
|
|
|
|
| 12 |
The model is trained based on ResNet18 [1] with the last fully connected layer replaced by a 1x1 convolution layer.
|
| 13 |

|
| 14 |
|
|
|
|
| 25 |
- Modality: Histopathology
|
| 26 |
- Size: 270 WSIs for training/validation, 48 WSIs for testing
|
| 27 |
|
| 28 |
+
### Preprocessing
|
| 29 |
|
| 30 |
This bundle expects the training/validation data (whole slide images) reside in a `{data_root}/training/images`. By default `data_root` is pointing to `/workspace/data/medical/pathology/` You can modify `data_root` in the bundle config files to point to a different directory.
|
| 31 |
|
|
|
|
| 33 |
|
| 34 |
Please refer to "Annotation" section of [Camelyon challenge](https://camelyon17.grand-challenge.org/Data/) to prepare ground truth images, which are needed for FROC computation. By default, this data set is expected to be at `/workspace/data/medical/pathology/ground_truths`. But it can be modified in `evaluate_froc.sh`.
|
| 35 |
|
| 36 |
+
## Training configuration
|
| 37 |
|
| 38 |
The training was performed with the following:
|
| 39 |
|
|
|
|
| 46 |
- Loss: BCEWithLogitsLoss
|
| 47 |
- Whole slide image reader: cuCIM (if running on Windows or Mac, please install `OpenSlide` on your system and change `wsi_reader` to "OpenSlide")
|
| 48 |
|
| 49 |
+
### Input
|
|
|
|
|
|
|
| 50 |
|
| 51 |
+
The training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 52 |
|
| 53 |
+
### Output
|
| 54 |
|
| 55 |
+
A probability number of the input patch being tumor or normal.
|
| 56 |
|
| 57 |
+
### Inference on a WSI
|
| 58 |
|
| 59 |
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 60 |
|
| 61 |
+
## Performance
|
| 62 |
|
| 63 |
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 64 |
This model achieve the ~0.91 accuracy on validation patches, and FROC of 0.685 on the 48 Camelyon testing data that have ground truth annotations available.
|
|
|
|
| 65 |
|
| 66 |
+

|
| 67 |
+
|
| 68 |
+
## MONAI Bundle Commands
|
| 69 |
+
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
|
| 70 |
+
|
| 71 |
+
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
|
| 72 |
|
| 73 |
+
#### Execute training:
|
| 74 |
|
| 75 |
```
|
| 76 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 77 |
```
|
| 78 |
|
| 79 |
+
#### Override the `train` config to execute multi-GPU training:
|
| 80 |
|
| 81 |
```
|
| 82 |
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 83 |
```
|
| 84 |
|
| 85 |
+
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html).
|
|
|
|
| 86 |
|
| 87 |
+
#### Execute inference:
|
| 88 |
|
| 89 |
```
|
| 90 |
CUDA_LAUNCH_BLOCKING=1 python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 91 |
```
|
| 92 |
|
| 93 |
+
#### Evaluate FROC metric:
|
| 94 |
|
| 95 |
```
|
| 96 |
cd scripts && source evaluate_froc.sh
|
| 97 |
```
|
| 98 |
|
| 99 |
+
#### Export checkpoint to TorchScript file:
|
| 100 |
|
| 101 |
TorchScript conversion is currently not supported.
|
| 102 |
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.4.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.4.3": "fix wrong figure url",
|
| 6 |
"0.4.2": "update metadata with new metrics",
|
| 7 |
"0.4.1": "Fix inference print logger and froc",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.4.4",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.4.4": "restructure readme to match updated template",
|
| 6 |
"0.4.3": "fix wrong figure url",
|
| 7 |
"0.4.2": "update metadata with new metrics",
|
| 8 |
"0.4.1": "Fix inference print logger and froc",
|
docs/README.md
CHANGED
|
@@ -2,8 +2,6 @@
|
|
| 2 |
|
| 3 |
A pre-trained model for automated detection of metastases in whole-slide histopathology images.
|
| 4 |
|
| 5 |
-
## Workflow
|
| 6 |
-
|
| 7 |
The model is trained based on ResNet18 [1] with the last fully connected layer replaced by a 1x1 convolution layer.
|
| 8 |

|
| 9 |
|
|
@@ -20,7 +18,7 @@ Annotation information are adopted from [NCRF/jsons](https://github.com/baidu-re
|
|
| 20 |
- Modality: Histopathology
|
| 21 |
- Size: 270 WSIs for training/validation, 48 WSIs for testing
|
| 22 |
|
| 23 |
-
###
|
| 24 |
|
| 25 |
This bundle expects the training/validation data (whole slide images) reside in a `{data_root}/training/images`. By default `data_root` is pointing to `/workspace/data/medical/pathology/` You can modify `data_root` in the bundle config files to point to a different directory.
|
| 26 |
|
|
@@ -28,7 +26,7 @@ To reduce the computation burden during the inference, patches are extracted onl
|
|
| 28 |
|
| 29 |
Please refer to "Annotation" section of [Camelyon challenge](https://camelyon17.grand-challenge.org/Data/) to prepare ground truth images, which are needed for FROC computation. By default, this data set is expected to be at `/workspace/data/medical/pathology/ground_truths`. But it can be modified in `evaluate_froc.sh`.
|
| 30 |
|
| 31 |
-
|
| 32 |
|
| 33 |
The training was performed with the following:
|
| 34 |
|
|
@@ -41,61 +39,57 @@ The training was performed with the following:
|
|
| 41 |
- Loss: BCEWithLogitsLoss
|
| 42 |
- Whole slide image reader: cuCIM (if running on Windows or Mac, please install `OpenSlide` on your system and change `wsi_reader` to "OpenSlide")
|
| 43 |
|
| 44 |
-
|
| 45 |
-
|
| 46 |
-
Input: Input for the training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
| 47 |
|
| 48 |
-
|
| 49 |
-
2. Randomly applying color jittering
|
| 50 |
-
3. Randomly applying spatial flipping
|
| 51 |
-
4. Randomly applying spatial rotation
|
| 52 |
-
5. Randomly applying spatial zooming
|
| 53 |
-
6. Randomly applying intensity scaling
|
| 54 |
|
| 55 |
-
|
| 56 |
|
| 57 |
-
|
| 58 |
|
| 59 |
-
|
| 60 |
|
| 61 |
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 62 |
|
| 63 |
-
|
| 64 |
|
| 65 |
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 66 |
This model achieve the ~0.91 accuracy on validation patches, and FROC of 0.685 on the 48 Camelyon testing data that have ground truth annotations available.
|
| 67 |
-

|
| 68 |
|
| 69 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 70 |
|
| 71 |
-
Execute training:
|
| 72 |
|
| 73 |
```
|
| 74 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 75 |
```
|
| 76 |
|
| 77 |
-
Override the `train` config to execute multi-GPU training:
|
| 78 |
|
| 79 |
```
|
| 80 |
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 81 |
```
|
| 82 |
|
| 83 |
-
Please note that the distributed training
|
| 84 |
-
Please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html) for more details.
|
| 85 |
|
| 86 |
-
Execute inference:
|
| 87 |
|
| 88 |
```
|
| 89 |
CUDA_LAUNCH_BLOCKING=1 python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 90 |
```
|
| 91 |
|
| 92 |
-
Evaluate FROC metric:
|
| 93 |
|
| 94 |
```
|
| 95 |
cd scripts && source evaluate_froc.sh
|
| 96 |
```
|
| 97 |
|
| 98 |
-
Export checkpoint to TorchScript file:
|
| 99 |
|
| 100 |
TorchScript conversion is currently not supported.
|
| 101 |
|
|
|
|
| 2 |
|
| 3 |
A pre-trained model for automated detection of metastases in whole-slide histopathology images.
|
| 4 |
|
|
|
|
|
|
|
| 5 |
The model is trained based on ResNet18 [1] with the last fully connected layer replaced by a 1x1 convolution layer.
|
| 6 |

|
| 7 |
|
|
|
|
| 18 |
- Modality: Histopathology
|
| 19 |
- Size: 270 WSIs for training/validation, 48 WSIs for testing
|
| 20 |
|
| 21 |
+
### Preprocessing
|
| 22 |
|
| 23 |
This bundle expects the training/validation data (whole slide images) reside in a `{data_root}/training/images`. By default `data_root` is pointing to `/workspace/data/medical/pathology/` You can modify `data_root` in the bundle config files to point to a different directory.
|
| 24 |
|
|
|
|
| 26 |
|
| 27 |
Please refer to "Annotation" section of [Camelyon challenge](https://camelyon17.grand-challenge.org/Data/) to prepare ground truth images, which are needed for FROC computation. By default, this data set is expected to be at `/workspace/data/medical/pathology/ground_truths`. But it can be modified in `evaluate_froc.sh`.
|
| 28 |
|
| 29 |
+
## Training configuration
|
| 30 |
|
| 31 |
The training was performed with the following:
|
| 32 |
|
|
|
|
| 39 |
- Loss: BCEWithLogitsLoss
|
| 40 |
- Whole slide image reader: cuCIM (if running on Windows or Mac, please install `OpenSlide` on your system and change `wsi_reader` to "OpenSlide")
|
| 41 |
|
| 42 |
+
### Input
|
|
|
|
|
|
|
| 43 |
|
| 44 |
+
The training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 45 |
|
| 46 |
+
### Output
|
| 47 |
|
| 48 |
+
A probability number of the input patch being tumor or normal.
|
| 49 |
|
| 50 |
+
### Inference on a WSI
|
| 51 |
|
| 52 |
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 53 |
|
| 54 |
+
## Performance
|
| 55 |
|
| 56 |
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 57 |
This model achieve the ~0.91 accuracy on validation patches, and FROC of 0.685 on the 48 Camelyon testing data that have ground truth annotations available.
|
|
|
|
| 58 |
|
| 59 |
+

|
| 60 |
+
|
| 61 |
+
## MONAI Bundle Commands
|
| 62 |
+
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
|
| 63 |
+
|
| 64 |
+
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
|
| 65 |
|
| 66 |
+
#### Execute training:
|
| 67 |
|
| 68 |
```
|
| 69 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 70 |
```
|
| 71 |
|
| 72 |
+
#### Override the `train` config to execute multi-GPU training:
|
| 73 |
|
| 74 |
```
|
| 75 |
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 76 |
```
|
| 77 |
|
| 78 |
+
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html).
|
|
|
|
| 79 |
|
| 80 |
+
#### Execute inference:
|
| 81 |
|
| 82 |
```
|
| 83 |
CUDA_LAUNCH_BLOCKING=1 python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 84 |
```
|
| 85 |
|
| 86 |
+
#### Evaluate FROC metric:
|
| 87 |
|
| 88 |
```
|
| 89 |
cd scripts && source evaluate_froc.sh
|
| 90 |
```
|
| 91 |
|
| 92 |
+
#### Export checkpoint to TorchScript file:
|
| 93 |
|
| 94 |
TorchScript conversion is currently not supported.
|
| 95 |
|