Spaces:
Running
Running
Upload folder using huggingface_hub
Browse files- .gitattributes +1 -0
- .github/workflows/update_space.yml +28 -0
- .gitignore +168 -0
- .gradio/cached_examples/19/Heart Lungs/b7614a6e0559aa743534/image.webp +0 -0
- .gradio/cached_examples/19/Pneumonia/693f3f20ddebe6f56bfa/image.webp +0 -0
- .gradio/cached_examples/19/Pneumothorax/74fe29695b6499d59559/image.webp +0 -0
- .gradio/cached_examples/19/log.csv +5 -0
- .gradio/certificate.pem +31 -0
- README.md +3 -9
- app.py +179 -0
- examples/cxr.png +3 -0
- requirements.txt +8 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
examples/cxr.png filter=lfs diff=lfs merge=lfs -text
|
.github/workflows/update_space.yml
ADDED
|
@@ -0,0 +1,28 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
name: Run Python script
|
| 2 |
+
|
| 3 |
+
on:
|
| 4 |
+
push:
|
| 5 |
+
branches:
|
| 6 |
+
- main
|
| 7 |
+
|
| 8 |
+
jobs:
|
| 9 |
+
build:
|
| 10 |
+
runs-on: ubuntu-latest
|
| 11 |
+
|
| 12 |
+
steps:
|
| 13 |
+
- name: Checkout
|
| 14 |
+
uses: actions/checkout@v2
|
| 15 |
+
|
| 16 |
+
- name: Set up Python
|
| 17 |
+
uses: actions/setup-python@v2
|
| 18 |
+
with:
|
| 19 |
+
python-version: '3.9'
|
| 20 |
+
|
| 21 |
+
- name: Install Gradio
|
| 22 |
+
run: python -m pip install gradio
|
| 23 |
+
|
| 24 |
+
- name: Log in to Hugging Face
|
| 25 |
+
run: python -c 'import huggingface_hub; huggingface_hub.login(token="${{ secrets.hf_token }}")'
|
| 26 |
+
|
| 27 |
+
- name: Deploy to Spaces
|
| 28 |
+
run: gradio deploy
|
.gitignore
ADDED
|
@@ -0,0 +1,168 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Byte-compiled / optimized / DLL files
|
| 2 |
+
__pycache__/
|
| 3 |
+
*.py[cod]
|
| 4 |
+
*$py.class
|
| 5 |
+
|
| 6 |
+
# C extensions
|
| 7 |
+
*.so
|
| 8 |
+
|
| 9 |
+
# Distribution / packaging
|
| 10 |
+
.Python
|
| 11 |
+
build/
|
| 12 |
+
develop-eggs/
|
| 13 |
+
dist/
|
| 14 |
+
downloads/
|
| 15 |
+
eggs/
|
| 16 |
+
.eggs/
|
| 17 |
+
lib/
|
| 18 |
+
lib64/
|
| 19 |
+
parts/
|
| 20 |
+
sdist/
|
| 21 |
+
var/
|
| 22 |
+
wheels/
|
| 23 |
+
share/python-wheels/
|
| 24 |
+
*.egg-info/
|
| 25 |
+
.installed.cfg
|
| 26 |
+
*.egg
|
| 27 |
+
MANIFEST
|
| 28 |
+
|
| 29 |
+
# PyInstaller
|
| 30 |
+
# Usually these files are written by a python script from a template
|
| 31 |
+
# before PyInstaller builds the exe, so as to inject date/other infos into it.
|
| 32 |
+
*.manifest
|
| 33 |
+
*.spec
|
| 34 |
+
|
| 35 |
+
# Installer logs
|
| 36 |
+
pip-log.txt
|
| 37 |
+
pip-delete-this-directory.txt
|
| 38 |
+
|
| 39 |
+
# Unit test / coverage reports
|
| 40 |
+
htmlcov/
|
| 41 |
+
.tox/
|
| 42 |
+
.nox/
|
| 43 |
+
.coverage
|
| 44 |
+
.coverage.*
|
| 45 |
+
.cache
|
| 46 |
+
nosetests.xml
|
| 47 |
+
coverage.xml
|
| 48 |
+
*.cover
|
| 49 |
+
*.py,cover
|
| 50 |
+
.hypothesis/
|
| 51 |
+
.pytest_cache/
|
| 52 |
+
cover/
|
| 53 |
+
|
| 54 |
+
# Translations
|
| 55 |
+
*.mo
|
| 56 |
+
*.pot
|
| 57 |
+
|
| 58 |
+
# Django stuff:
|
| 59 |
+
*.log
|
| 60 |
+
local_settings.py
|
| 61 |
+
db.sqlite3
|
| 62 |
+
db.sqlite3-journal
|
| 63 |
+
|
| 64 |
+
# Flask stuff:
|
| 65 |
+
instance/
|
| 66 |
+
.webassets-cache
|
| 67 |
+
|
| 68 |
+
# Scrapy stuff:
|
| 69 |
+
.scrapy
|
| 70 |
+
|
| 71 |
+
# Sphinx documentation
|
| 72 |
+
docs/_build/
|
| 73 |
+
|
| 74 |
+
# PyBuilder
|
| 75 |
+
.pybuilder/
|
| 76 |
+
target/
|
| 77 |
+
|
| 78 |
+
# Jupyter Notebook
|
| 79 |
+
.ipynb_checkpoints
|
| 80 |
+
|
| 81 |
+
# IPython
|
| 82 |
+
profile_default/
|
| 83 |
+
ipython_config.py
|
| 84 |
+
|
| 85 |
+
# pyenv
|
| 86 |
+
# For a library or package, you might want to ignore these files since the code is
|
| 87 |
+
# intended to run in multiple environments; otherwise, check them in:
|
| 88 |
+
# .python-version
|
| 89 |
+
|
| 90 |
+
# pipenv
|
| 91 |
+
# According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
|
| 92 |
+
# However, in case of collaboration, if having platform-specific dependencies or dependencies
|
| 93 |
+
# having no cross-platform support, pipenv may install dependencies that don't work, or not
|
| 94 |
+
# install all needed dependencies.
|
| 95 |
+
#Pipfile.lock
|
| 96 |
+
|
| 97 |
+
# UV
|
| 98 |
+
# Similar to Pipfile.lock, it is generally recommended to include uv.lock in version control.
|
| 99 |
+
# This is especially recommended for binary packages to ensure reproducibility, and is more
|
| 100 |
+
# commonly ignored for libraries.
|
| 101 |
+
#uv.lock
|
| 102 |
+
|
| 103 |
+
# poetry
|
| 104 |
+
# Similar to Pipfile.lock, it is generally recommended to include poetry.lock in version control.
|
| 105 |
+
# This is especially recommended for binary packages to ensure reproducibility, and is more
|
| 106 |
+
# commonly ignored for libraries.
|
| 107 |
+
# https://python-poetry.org/docs/basic-usage/#commit-your-poetrylock-file-to-version-control
|
| 108 |
+
#poetry.lock
|
| 109 |
+
|
| 110 |
+
# pdm
|
| 111 |
+
# Similar to Pipfile.lock, it is generally recommended to include pdm.lock in version control.
|
| 112 |
+
#pdm.lock
|
| 113 |
+
# pdm stores project-wide configurations in .pdm.toml, but it is recommended to not include it
|
| 114 |
+
# in version control.
|
| 115 |
+
# https://pdm.fming.dev/latest/usage/project/#working-with-version-control
|
| 116 |
+
.pdm.toml
|
| 117 |
+
.pdm-python
|
| 118 |
+
.pdm-build/
|
| 119 |
+
|
| 120 |
+
# PEP 582; used by e.g. github.com/David-OConnor/pyflow and github.com/pdm-project/pdm
|
| 121 |
+
__pypackages__/
|
| 122 |
+
|
| 123 |
+
# Celery stuff
|
| 124 |
+
celerybeat-schedule
|
| 125 |
+
celerybeat.pid
|
| 126 |
+
|
| 127 |
+
# SageMath parsed files
|
| 128 |
+
*.sage.py
|
| 129 |
+
|
| 130 |
+
# Environments
|
| 131 |
+
.env
|
| 132 |
+
.venv
|
| 133 |
+
env/
|
| 134 |
+
venv/
|
| 135 |
+
ENV/
|
| 136 |
+
env.bak/
|
| 137 |
+
venv.bak/
|
| 138 |
+
|
| 139 |
+
# Spyder project settings
|
| 140 |
+
.spyderproject
|
| 141 |
+
.spyproject
|
| 142 |
+
|
| 143 |
+
# Rope project settings
|
| 144 |
+
.ropeproject
|
| 145 |
+
|
| 146 |
+
# mkdocs documentation
|
| 147 |
+
/site
|
| 148 |
+
|
| 149 |
+
# mypy
|
| 150 |
+
.mypy_cache/
|
| 151 |
+
.dmypy.json
|
| 152 |
+
dmypy.json
|
| 153 |
+
|
| 154 |
+
# Pyre type checker
|
| 155 |
+
.pyre/
|
| 156 |
+
|
| 157 |
+
# pytype static type analyzer
|
| 158 |
+
.pytype/
|
| 159 |
+
|
| 160 |
+
# Cython debug symbols
|
| 161 |
+
cython_debug/
|
| 162 |
+
|
| 163 |
+
# PyCharm
|
| 164 |
+
# JetBrains specific template is maintained in a separate JetBrains.gitignore that can
|
| 165 |
+
# be found at https://github.com/github/gitignore/blob/main/Global/JetBrains.gitignore
|
| 166 |
+
# and can be added to the global gitignore or merged into this file. For a more nuclear
|
| 167 |
+
# option (not recommended) you can uncomment the following to ignore the entire idea folder.
|
| 168 |
+
#.idea/
|
.gradio/cached_examples/19/Heart Lungs/b7614a6e0559aa743534/image.webp
ADDED

|
.gradio/cached_examples/19/Pneumonia/693f3f20ddebe6f56bfa/image.webp
ADDED
|
.gradio/cached_examples/19/Pneumothorax/74fe29695b6499d59559/image.webp
ADDED

|
.gradio/cached_examples/19/log.csv
ADDED
|
@@ -0,0 +1,5 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
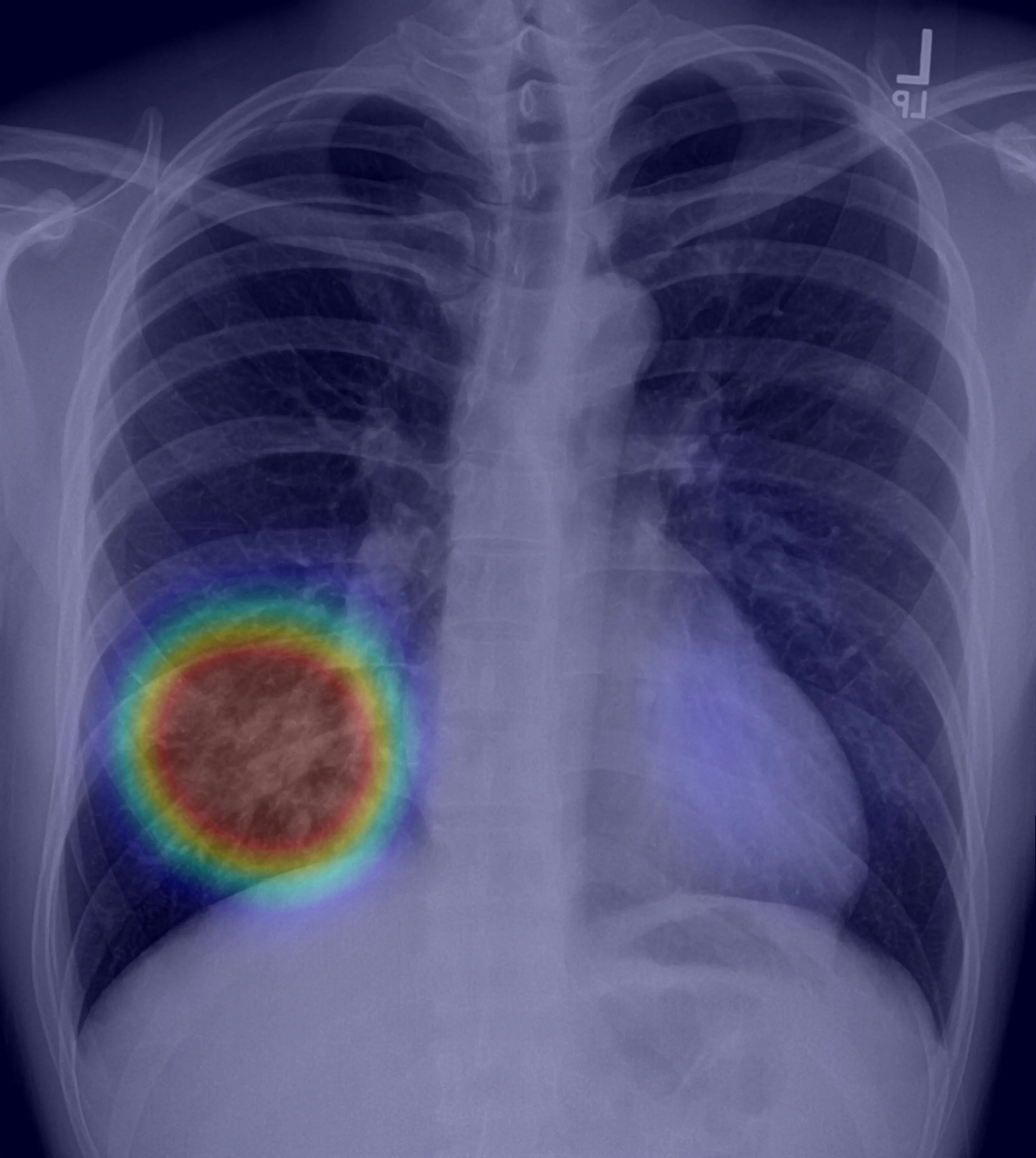
output 0,output 1,Heart & Lungs,Pneumonia,Pneumothorax,timestamp
|
| 2 |
+
"This is a frontal chest radiograph (PA projection).
|
| 3 |
+
The patient's predicted age is 37 years.
|
| 4 |
+
The patient's predicted sex is male.
|
| 5 |
+
The estimated cardiothoracic radio (CTR) is 0.50.","{""label"": ""Pneumonia"", ""confidences"": [{""label"": ""Pneumonia"", ""confidence"": 0.8377197980880737}, {""label"": ""Pneumothorax"", ""confidence"": 0.0041968440636992455}]}","{""path"": "".gradio/cached_examples/19/Heart Lungs/b7614a6e0559aa743534/image.webp"", ""url"": ""/gradio_api/file=/tmp/gradio/0ab95a795cbdce59306466d927e2266137153c344fa7839014c8bbf3aafc0258/image.webp"", ""size"": null, ""orig_name"": ""image.webp"", ""mime_type"": null, ""is_stream"": false, ""meta"": {""_type"": ""gradio.FileData""}}","{""path"": "".gradio/cached_examples/19/Pneumonia/693f3f20ddebe6f56bfa/image.webp"", ""url"": ""/gradio_api/file=/tmp/gradio/e22234adf5bd8f48a90fe6f5cb28c9e4d9ab4b9d533750776c1845ee08daaf15/image.webp"", ""size"": null, ""orig_name"": ""image.webp"", ""mime_type"": null, ""is_stream"": false, ""meta"": {""_type"": ""gradio.FileData""}}","{""path"": "".gradio/cached_examples/19/Pneumothorax/74fe29695b6499d59559/image.webp"", ""url"": ""/gradio_api/file=/tmp/gradio/09544f2f42aca03303abcd3386bc452d8819f9dc2f823f0a8813cf6e209eb20e/image.webp"", ""size"": null, ""orig_name"": ""image.webp"", ""mime_type"": null, ""is_stream"": false, ""meta"": {""_type"": ""gradio.FileData""}}",2024-12-27 12:16:10.290917
|
.gradio/certificate.pem
ADDED
|
@@ -0,0 +1,31 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
-----BEGIN CERTIFICATE-----
|
| 2 |
+
MIIFazCCA1OgAwIBAgIRAIIQz7DSQONZRGPgu2OCiwAwDQYJKoZIhvcNAQELBQAw
|
| 3 |
+
TzELMAkGA1UEBhMCVVMxKTAnBgNVBAoTIEludGVybmV0IFNlY3VyaXR5IFJlc2Vh
|
| 4 |
+
cmNoIEdyb3VwMRUwEwYDVQQDEwxJU1JHIFJvb3QgWDEwHhcNMTUwNjA0MTEwNDM4
|
| 5 |
+
WhcNMzUwNjA0MTEwNDM4WjBPMQswCQYDVQQGEwJVUzEpMCcGA1UEChMgSW50ZXJu
|
| 6 |
+
ZXQgU2VjdXJpdHkgUmVzZWFyY2ggR3JvdXAxFTATBgNVBAMTDElTUkcgUm9vdCBY
|
| 7 |
+
MTCCAiIwDQYJKoZIhvcNAQEBBQADggIPADCCAgoCggIBAK3oJHP0FDfzm54rVygc
|
| 8 |
+
h77ct984kIxuPOZXoHj3dcKi/vVqbvYATyjb3miGbESTtrFj/RQSa78f0uoxmyF+
|
| 9 |
+
0TM8ukj13Xnfs7j/EvEhmkvBioZxaUpmZmyPfjxwv60pIgbz5MDmgK7iS4+3mX6U
|
| 10 |
+
A5/TR5d8mUgjU+g4rk8Kb4Mu0UlXjIB0ttov0DiNewNwIRt18jA8+o+u3dpjq+sW
|
| 11 |
+
T8KOEUt+zwvo/7V3LvSye0rgTBIlDHCNAymg4VMk7BPZ7hm/ELNKjD+Jo2FR3qyH
|
| 12 |
+
B5T0Y3HsLuJvW5iB4YlcNHlsdu87kGJ55tukmi8mxdAQ4Q7e2RCOFvu396j3x+UC
|
| 13 |
+
B5iPNgiV5+I3lg02dZ77DnKxHZu8A/lJBdiB3QW0KtZB6awBdpUKD9jf1b0SHzUv
|
| 14 |
+
KBds0pjBqAlkd25HN7rOrFleaJ1/ctaJxQZBKT5ZPt0m9STJEadao0xAH0ahmbWn
|
| 15 |
+
OlFuhjuefXKnEgV4We0+UXgVCwOPjdAvBbI+e0ocS3MFEvzG6uBQE3xDk3SzynTn
|
| 16 |
+
jh8BCNAw1FtxNrQHusEwMFxIt4I7mKZ9YIqioymCzLq9gwQbooMDQaHWBfEbwrbw
|
| 17 |
+
qHyGO0aoSCqI3Haadr8faqU9GY/rOPNk3sgrDQoo//fb4hVC1CLQJ13hef4Y53CI
|
| 18 |
+
rU7m2Ys6xt0nUW7/vGT1M0NPAgMBAAGjQjBAMA4GA1UdDwEB/wQEAwIBBjAPBgNV
|
| 19 |
+
HRMBAf8EBTADAQH/MB0GA1UdDgQWBBR5tFnme7bl5AFzgAiIyBpY9umbbjANBgkq
|
| 20 |
+
hkiG9w0BAQsFAAOCAgEAVR9YqbyyqFDQDLHYGmkgJykIrGF1XIpu+ILlaS/V9lZL
|
| 21 |
+
ubhzEFnTIZd+50xx+7LSYK05qAvqFyFWhfFQDlnrzuBZ6brJFe+GnY+EgPbk6ZGQ
|
| 22 |
+
3BebYhtF8GaV0nxvwuo77x/Py9auJ/GpsMiu/X1+mvoiBOv/2X/qkSsisRcOj/KK
|
| 23 |
+
NFtY2PwByVS5uCbMiogziUwthDyC3+6WVwW6LLv3xLfHTjuCvjHIInNzktHCgKQ5
|
| 24 |
+
ORAzI4JMPJ+GslWYHb4phowim57iaztXOoJwTdwJx4nLCgdNbOhdjsnvzqvHu7Ur
|
| 25 |
+
TkXWStAmzOVyyghqpZXjFaH3pO3JLF+l+/+sKAIuvtd7u+Nxe5AW0wdeRlN8NwdC
|
| 26 |
+
jNPElpzVmbUq4JUagEiuTDkHzsxHpFKVK7q4+63SM1N95R1NbdWhscdCb+ZAJzVc
|
| 27 |
+
oyi3B43njTOQ5yOf+1CceWxG1bQVs5ZufpsMljq4Ui0/1lvh+wjChP4kqKOJ2qxq
|
| 28 |
+
4RgqsahDYVvTH9w7jXbyLeiNdd8XM2w9U/t7y0Ff/9yi0GE44Za4rF2LN9d11TPA
|
| 29 |
+
mRGunUHBcnWEvgJBQl9nJEiU0Zsnvgc/ubhPgXRR4Xq37Z0j4r7g1SgEEzwxA57d
|
| 30 |
+
emyPxgcYxn/eR44/KJ4EBs+lVDR3veyJm+kXQ99b21/+jh5Xos1AnX5iItreGCc=
|
| 31 |
+
-----END CERTIFICATE-----
|
README.md
CHANGED
|
@@ -1,12 +1,6 @@
|
|
| 1 |
---
|
| 2 |
-
title:
|
| 3 |
-
emoji: 🏆
|
| 4 |
-
colorFrom: green
|
| 5 |
-
colorTo: blue
|
| 6 |
-
sdk: gradio
|
| 7 |
-
sdk_version: 5.9.1
|
| 8 |
app_file: app.py
|
| 9 |
-
|
|
|
|
| 10 |
---
|
| 11 |
-
|
| 12 |
-
Check out the configuration reference at https://huggingface.co/docs/hub/spaces-config-reference
|
|
|
|
| 1 |
---
|
| 2 |
+
title: chest-x-ray-ai
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 3 |
app_file: app.py
|
| 4 |
+
sdk: gradio
|
| 5 |
+
sdk_version: 5.9.0
|
| 6 |
---
|
|
|
|
|
|
app.py
ADDED
|
@@ -0,0 +1,179 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import cv2
|
| 2 |
+
import gradio as gr
|
| 3 |
+
import numpy as np
|
| 4 |
+
import spaces
|
| 5 |
+
import torch
|
| 6 |
+
import torch.nn.functional as F
|
| 7 |
+
|
| 8 |
+
from einops import rearrange
|
| 9 |
+
|
| 10 |
+
from transformers import AutoModel
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
def calculate_ctr(mask: np.ndarray) -> float:
|
| 14 |
+
# mask.ndim = 2, (height, width)
|
| 15 |
+
lungs = np.zeros_like(mask)
|
| 16 |
+
lungs[mask == 1] = 1
|
| 17 |
+
lungs[mask == 2] = 1
|
| 18 |
+
heart = (mask == 3).astype("int")
|
| 19 |
+
y, x = np.stack(np.where(lungs == 1))
|
| 20 |
+
lung_min = x.min()
|
| 21 |
+
lung_max = x.max()
|
| 22 |
+
y, x = np.stack(np.where(heart == 1))
|
| 23 |
+
heart_min = x.min()
|
| 24 |
+
heart_max = x.max()
|
| 25 |
+
lung_range = lung_max - lung_min
|
| 26 |
+
heart_range = heart_max - heart_min
|
| 27 |
+
return heart_range / lung_range
|
| 28 |
+
|
| 29 |
+
|
| 30 |
+
def make_overlay(
|
| 31 |
+
img: np.ndarray, mask: np.ndarray, alpha: float = 0.7
|
| 32 |
+
) -> np.ndarray[np.uint8]:
|
| 33 |
+
overlay = alpha * img + (1 - alpha) * mask
|
| 34 |
+
return overlay.astype(np.uint8)
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
@spaces.GPU
|
| 38 |
+
def predict(Radiograph):
|
| 39 |
+
rg = cv2.cvtColor(Radiograph, cv2.COLOR_GRAY2RGB)
|
| 40 |
+
x = cxr_info_model.preprocess(Radiograph)
|
| 41 |
+
x = torch.from_numpy(x).float().to(device)
|
| 42 |
+
x = rearrange(x, "h w -> 1 1 h w")
|
| 43 |
+
|
| 44 |
+
with torch.inference_mode():
|
| 45 |
+
info_out = cxr_info_model(x)
|
| 46 |
+
|
| 47 |
+
info_mask = info_out["mask"]
|
| 48 |
+
h, w = rg.shape[:2]
|
| 49 |
+
info_mask = F.interpolate(info_mask, size=(h, w), mode="bilinear")
|
| 50 |
+
info_mask = info_mask.argmax(1)[0]
|
| 51 |
+
info_mask_3ch = F.one_hot(info_mask, num_classes=4)[..., 1:]
|
| 52 |
+
info_mask_3ch = (info_mask_3ch.cpu().numpy() * 255).astype(np.uint8)
|
| 53 |
+
info_overlay = make_overlay(rg, info_mask_3ch[..., ::-1])
|
| 54 |
+
|
| 55 |
+
view = info_out["view"].argmax(1).item()
|
| 56 |
+
info_string = ""
|
| 57 |
+
if view in {0, 1}:
|
| 58 |
+
info_string += "This is a frontal chest radiograph "
|
| 59 |
+
if view == 0:
|
| 60 |
+
info_string += "(AP projection)."
|
| 61 |
+
elif view == 1:
|
| 62 |
+
info_string += "(PA projection)."
|
| 63 |
+
elif view == 2:
|
| 64 |
+
info_string += "This is a lateral chest radiograph."
|
| 65 |
+
|
| 66 |
+
age = info_out["age"].item()
|
| 67 |
+
info_string += f"\nThe patient's predicted age is {round(age)} years."
|
| 68 |
+
sex = info_out["female"].item()
|
| 69 |
+
if sex < 0.5:
|
| 70 |
+
sex = "male"
|
| 71 |
+
else:
|
| 72 |
+
sex = "female"
|
| 73 |
+
info_string += f"\nThe patient's predicted sex is {sex}."
|
| 74 |
+
|
| 75 |
+
if view in {0, 1}:
|
| 76 |
+
ctr = calculate_ctr(info_mask.cpu().numpy())
|
| 77 |
+
info_string += f"\nThe estimated cardiothoracic radio (CTR) is {ctr:0.2f}."
|
| 78 |
+
if view == 0:
|
| 79 |
+
info_string += (
|
| 80 |
+
"\nNote that the cardiac silhuoette is magnified in the AP projection."
|
| 81 |
+
)
|
| 82 |
+
|
| 83 |
+
if view == 2:
|
| 84 |
+
info_string += (
|
| 85 |
+
"\nNOTE: The below outputs are NOT VALID for lateral radiographs."
|
| 86 |
+
)
|
| 87 |
+
|
| 88 |
+
x = pna_model.preprocess(Radiograph)
|
| 89 |
+
x = torch.from_numpy(x).float().to(device)
|
| 90 |
+
x = rearrange(x, "h w -> 1 1 h w")
|
| 91 |
+
|
| 92 |
+
with torch.inference_mode():
|
| 93 |
+
pna_out = pna_model(x)
|
| 94 |
+
|
| 95 |
+
pna_mask = pna_out["mask"]
|
| 96 |
+
h, w = rg.shape[:2]
|
| 97 |
+
pna_mask = F.interpolate(pna_mask, size=(h, w), mode="bilinear")
|
| 98 |
+
pna_mask = (pna_mask.cpu().numpy()[0, 0] * 255).astype(np.uint8)
|
| 99 |
+
pna_mask = cv2.applyColorMap(pna_mask, cv2.COLORMAP_JET)
|
| 100 |
+
pna_overlay = make_overlay(rg, pna_mask[..., ::-1])
|
| 101 |
+
|
| 102 |
+
x = ptx_model.preprocess(Radiograph)
|
| 103 |
+
x = torch.from_numpy(x).float().to(device)
|
| 104 |
+
x = rearrange(x, "h w -> 1 1 h w")
|
| 105 |
+
|
| 106 |
+
with torch.inference_mode():
|
| 107 |
+
ptx_out = ptx_model(x)
|
| 108 |
+
|
| 109 |
+
ptx_mask = ptx_out["mask"]
|
| 110 |
+
h, w = rg.shape[:2]
|
| 111 |
+
ptx_mask = F.interpolate(ptx_mask, size=(h, w), mode="bilinear")
|
| 112 |
+
ptx_mask = (ptx_mask.cpu().numpy()[0, 0] * 255).astype(np.uint8)
|
| 113 |
+
ptx_mask = cv2.applyColorMap(ptx_mask, cv2.COLORMAP_JET)
|
| 114 |
+
ptx_overlay = make_overlay(rg, ptx_mask[..., ::-1])
|
| 115 |
+
|
| 116 |
+
preds = {"Pneumonia": pna_out["cls"].item(), "Pneumothorax": ptx_out["cls"].item()}
|
| 117 |
+
return [info_string, preds, info_overlay, pna_overlay, ptx_overlay]
|
| 118 |
+
|
| 119 |
+
|
| 120 |
+
image = gr.Image(image_mode="L")
|
| 121 |
+
info_textbox = gr.Textbox(show_label=False)
|
| 122 |
+
labels = gr.Label(show_label=False, show_heading=False)
|
| 123 |
+
heatmap1 = gr.Image(image_mode="RGB", label="Heart & Lungs")
|
| 124 |
+
heatmap2 = gr.Image(image_mode="RGB", label="Pneumonia")
|
| 125 |
+
heatmap3 = gr.Image(image_mode="RGB", label="Pneumothorax")
|
| 126 |
+
|
| 127 |
+
with gr.Blocks() as demo:
|
| 128 |
+
gr.Markdown(
|
| 129 |
+
"""
|
| 130 |
+
# Deep Learning for Chest Radiographs
|
| 131 |
+
|
| 132 |
+
This demo uses 3 models for chest radiographs:
|
| 133 |
+
1) Heart and lungs segmentation, with age, view, and sex prediction <https://huggingface.co/ianpan/chest-x-ray-basic>
|
| 134 |
+
2) Pneumonia classification and segmentation <https://huggingface.co/ianpan/pneumonia-cxr>
|
| 135 |
+
3) Pneumothorax classification and segmentation <https://huggingface.co/ianpan/pneumothorax-cxr>
|
| 136 |
+
|
| 137 |
+
Note that the pneumonia and pneumothorax heatmaps produced by this model are based on pixel-level segmentation maps.
|
| 138 |
+
Thus, they are expected to be more accurate than non-explicit localization methods such as GradCAM.
|
| 139 |
+
|
| 140 |
+
The example radiograph is my own, from when I had pneumonia.
|
| 141 |
+
|
| 142 |
+
This model is for demonstration purposes only and has NOT been approved by any regulatory agency for clinical use. The user assumes
|
| 143 |
+
any and all responsibility regarding their own use of this model and its outputs. Do NOT upload any images containing protected
|
| 144 |
+
health information, as this demonstration is not compliant with patient privacy laws.
|
| 145 |
+
|
| 146 |
+
Created by: Ian Pan, <https://ianpan.me>
|
| 147 |
+
|
| 148 |
+
Last updated: December 27, 2024
|
| 149 |
+
"""
|
| 150 |
+
)
|
| 151 |
+
gr.Interface(
|
| 152 |
+
fn=predict,
|
| 153 |
+
inputs=image,
|
| 154 |
+
outputs=[info_textbox, labels, heatmap1, heatmap2, heatmap3],
|
| 155 |
+
examples=["examples/cxr.png"],
|
| 156 |
+
cache_examples=True,
|
| 157 |
+
)
|
| 158 |
+
|
| 159 |
+
if __name__ == "__main__":
|
| 160 |
+
device = "cuda" if torch.cuda.is_available() else "cpu"
|
| 161 |
+
print(f"Using device `{device}` ...")
|
| 162 |
+
|
| 163 |
+
cxr_info_model = (
|
| 164 |
+
AutoModel.from_pretrained("ianpan/chest-x-ray-basic", trust_remote_code=True)
|
| 165 |
+
.eval()
|
| 166 |
+
.to(device)
|
| 167 |
+
)
|
| 168 |
+
pna_model = (
|
| 169 |
+
AutoModel.from_pretrained("ianpan/pneumonia-cxr", trust_remote_code=True)
|
| 170 |
+
.eval()
|
| 171 |
+
.to(device)
|
| 172 |
+
)
|
| 173 |
+
ptx_model = (
|
| 174 |
+
AutoModel.from_pretrained("ianpan/pneumothorax-cxr", trust_remote_code=True)
|
| 175 |
+
.eval()
|
| 176 |
+
.to(device)
|
| 177 |
+
)
|
| 178 |
+
|
| 179 |
+
demo.launch(share=True)
|
examples/cxr.png
ADDED

|
Git LFS Details
|
requirements.txt
ADDED
|
@@ -0,0 +1,8 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
albumentations
|
| 2 |
+
einops
|
| 3 |
+
gradio
|
| 4 |
+
scikit-image
|
| 5 |
+
spaces
|
| 6 |
+
timm
|
| 7 |
+
torch
|
| 8 |
+
transformers
|